|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 17 |
c.7964A>G |
8192A>G |
p.Q2655R |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
0.529138898 |
1.24584768 |
- |
- |
- |
0.659226469 |
- |
| 17 |
c.7975A>G |
8203A>G |
p.R2659G |
0.999974994 |
5 - Pathogenic |
Parsons et al 2019 |
- |
0.81 |
0.34 |
0.02 |
0.81 |
1950 |
0.86115312 |
5.068240677 |
1.102157073 |
- |
9380.278069 |
- |
| 17 |
c.7976G>A |
8204G>A |
R2659K |
0.99943 |
5 - Definitely pathogenic |
Farrugia DJ et al., Cancer Research 68:3523-3531, 2008. |
Vallee MP et al., Human Mutation 37: 627-639, 2016 |
0.29 |
0.34 |
0.00 |
0.34 |
- |
- |
1.55E+03 |
2.19 |
- |
3.39E+03 |
- |
| 17 |
c.7976G>C |
8204G>C |
p.R2659T |
0.998 |
5 - Definitely pathogenic |
Chenevix-Trench, G. et al., Cancer Research 66: 2019-2027, 2006. |
Vallee MP et al., Human Mutation 37: 627-639, 2016 |
0.81 |
0.34 |
0.00 |
0.97 |
14.7 |
1 |
1 |
1 |
- |
14.7 |
- |
| 17 |
c.7976+1G>A |
IVS17+1G>A |
- |
0.999776288 |
5 - Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
0.7080605 |
- |
160.5676159 |
1.215721465 |
- |
138.217302 |
- |
| 17 |
c.7976+23C>T |
IVS17+23C>T |
- |
2.09232E-09 |
1 - Not pathogenic or of no clinical significance |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.02 |
- |
2.613378994 |
- |
- |
3.92304E-08 |
0.000000102524 |
- |
| 17 |
c.7977-15T>G |
IVS17-15T>G |
- |
- |
- |
Parsons et al 2019 |
- |
- |
0.04 |
0.02 |
0.04 |
- |
- |
1.082639742 |
1.024615386 |
- |
1.109289337 |
- |
| 17 |
c.7977-1G>C |
IVS17-1G>C |
- |
0.999999998 |
5 - Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
1984739.681 |
10.20129138 |
- |
- |
- |
20246907.8 |
- |
| 18 |
c.7985C>A |
8213C>A |
p.T2662K |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.565976745 |
1.102157073 |
- |
0.623795272 |
- |
| 18 |
c.7985C>T |
8213C>T |
p.T2662M |
0.06403194 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
1.0157 |
- |
2.07443061 |
1.049836689 |
- |
2.212005032 |
- |
| 18 |
c.7987G>A |
8215G>A |
p.E2663K |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
0.81 |
0.882012538 |
1.129287087 |
- |
0.806796749 |
- |
| 18 |
c.7988A>T |
8216A>T |
p.E2663V |
0.999 |
5 - Definitely pathogenic |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.30 |
0.00 |
0.81 |
138.04 |
- |
1.12 |
1.51 |
- |
233.45 |
- |
| 18 |
c.7992T>G |
8220T>G |
p.I2664M |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.133553295 |
1.024615386 |
- |
1.161456147 |
- |
| 18 |
c.7994A>G
(Reported 2 times) |
8222A>G |
p.D2665G |
0.0183 |
2 - Likely not pathogenic or of little clinical significance |
Chenevix-Trench, G. et al., Cancer Research 66: 2019-2027, 2006. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.01 |
0.00 |
0.81 |
0.0021 |
1.2 |
- |
1.733 |
- |
0.00437 |
- |
| 18 |
c.8009C>T
(Reported 2 times) |
8237C>T |
p.S2670L |
0.5002 |
3 - Uncertain |
Chenevix-Trench, G. et al., Cancer Research 66: 2019-2027, 2006. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.29 |
0.01 |
0.00 |
0.29 |
2 |
1 |
- |
1.225 |
- |
2.45 |
- |
| 18 |
c.8011G>T |
8239G>T |
p.A2671S |
- |
- |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
- |
- |
1.249955187 |
1.024615386 |
- |
1.280723316 |
- |
| 18 |
c.8014A>G |
8242A>G |
p.I2672V |
0.11658096 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.293141788 |
1.102157073 |
- |
0.323088295 |
- |
| 18 |
c.8023A>G |
8251A>G |
p.I2675V |
0.999651188 |
5 - Pathogenic |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.64 |
605.1396929 |
0.88 |
2.883498045 |
1.049836689 |
- |
1612.054499 |
- |
| 18 |
c.8027T>C |
8255T>C |
p.M2676T |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
1.15 |
0.832640877 |
1.157084926 |
- |
1.10795164 |
- |
| 18 |
c.8032A>G |
8260A>G |
p.R2678G |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
- |
0.785763889 |
1.024615386 |
- |
0.80510577 |
- |
| 18 |
c.8035G>T |
8263G>T |
p.D2679Y |
0.953053101 |
4 - Likely Pathogenic |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
3.5432 |
1.06 |
1.237417811 |
1.024615386 |
- |
4.761883522 |
- |
| 18 |
c.8036A>G |
8264A>G |
p.D2679G |
- |
- |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
- |
- |
0.579160422 |
1.102157073 |
- |
0.638325755 |
- |
| 18 |
c.8039A>G |
8267A>G |
p.D2680G |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.801601969 |
1.049836689 |
- |
0.841551156 |
- |
| 18 |
c.8042C>G
(Reported 2 times) |
8270C>G |
p.T2681R |
0.1206 |
3 - Uncertain |
Spearman AD et al., J Clin Oncol, 26:5393-5400, 2008. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.03 |
0.01 |
0.00 |
0.03 |
- |
0.892 |
4.97 |
- |
- |
4.43324 |
- |
| 18 |
c.8051A>G |
8279A>G |
p.K2684R |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.173388514 |
1.049836689 |
- |
1.231866312 |
- |
|
|
| DNA change |
|
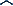
 | |
|
|
|
|
| Key Observational Reference |
 |
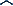
 | |
|
| Missense analysis Prior P |
 |
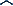
 | |
|
|
|
|
|
|
|
|
|
|
| 18 |
c.8057T>C |
8285T>C |
p.L2686P |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
- |
1.701629425 |
1.024615386 |
- |
1.743515689 |
- |
| 18 |
c.8084C>T |
8312C>T |
p.S2695L |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
1.77 |
0.456838357 |
1.129287087 |
- |
0.913145934 |
- |
| 18 |
c.8090G>A |
8318G>A |
p.S2697N |
0.001437459 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
0.040326 |
1.06 |
0.702884719 |
1.549157977 |
- |
0.046544763 |
- |
| 18 |
c.8113A>G |
8341A>G |
p.S2705G |
0.062461998 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
2.051897789 |
1.049836689 |
- |
2.15415758 |
- |
| 18 |
c.8117A>G |
8345A>G |
p.N2706S |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
0.69 |
1.229659475 |
1.405569136 |
- |
1.19257627 |
- |
| 18 |
c.8134G>A |
8362G>A |
p.D2712N |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.922253472 |
1.075678821 |
- |
0.992048528 |
- |
| 18 |
c.8135A>T |
8363A>T |
p.D2712V |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.066689585 |
1.102157073 |
- |
1.17565947 |
- |
| 18 |
c.8138C>A |
8366C>A |
p.T2713N |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.588608958 |
1.049836689 |
- |
1.667779968 |
- |
| 18 |
c.8141A>G |
8369A>G |
p.Q2714R |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.62046407 |
1.049836689 |
- |
0.651385944 |
- |
| 18 |
c.8149G>T |
8377G>T |
p.A2717S |
0.00000 |
1 - Not pathogenic or of no clinical significance |
Spurdle AB et al., J Clin Oncol, 26: 1657-1663, 2008. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.03 |
0.01 |
0.00 |
0.03 |
0.1211 |
1 |
- |
1.28E-03 |
- |
0.00016 |
- |
| 18 |
c.8156T>C |
8384T>C |
p.I2719T |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.508026915 |
1.024615386 |
- |
0.520532193 |
- |
| 18 |
c.8157T>G |
8385T>G |
p.I2719M |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.907743458 |
1.024615386 |
- |
0.930087913 |
- |
| 18 |
c.8162T>A |
8390T>A |
p.L2721H |
0.1752 |
3 - Uncertain |
Spearman AD et al., J Clin Oncol, 26:5393-5400, 2008. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.29 |
0.01 |
0.00 |
0.29 |
- |
- |
0.52 |
- |
- |
0.52 |
- |
| 18 |
c.8165C>G |
8393C>G |
p.T2722R |
0.9975 |
5 - Definitely pathogenic |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.01 |
0.00 |
0.81 |
10.715 |
- |
6.918 |
1.259 |
- |
93.3251 |
- |
| 18 |
c.8165C>T |
8393C>T |
p.T2722I |
0.929330881 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
- |
- |
2.798760809 |
1.102157073 |
- |
3.08467402 |
- |
| 18 |
c.8167G>C |
8395G>C |
p.D2723H |
1.0000 |
5 - Definitely pathogenic |
Goldgar et al., AJHG 75: 535-544, 2004. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.01 |
0.00 |
0.81 |
13731 |
- |
- |
2 |
- |
2.74620E+4 |
- |
| 18 |
c.8168A>G |
8396A>G |
P.D2723G |
0.998 |
5 - Definitely pathogenic |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Vallee MP et al., Human Mutation 37: 627-639, 2016 |
0.81 |
0.64 |
0.00 |
0.81 |
1 |
1 |
75.86 |
1.59 |
- |
120.6 |
- |
| 18 |
c.8182G>A |
8410G>A |
p.V2728I |
0.000345012 |
1 - Not pathogenic or of no clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
7.607259039 |
0.001466919 |
- |
- |
- |
0.011159236 |
- |
| 18 |
c.8187G>T |
8415G>T |
p.K2729N |
0.00005 |
1 - Not pathogenic or of no clinical significance |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.03 |
0.01 |
0.00 |
0.03 |
0.04169 |
- |
0.00617 |
6.30957 |
- |
0.00162 |
- |
| 18 |
c.8191C>G |
8419C>G |
p.Q2731E |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.953713744 |
1.024615386 |
- |
0.977189775 |
- |
| 18 |
c.8206C>G |
8434C>G |
p.L2736V |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.500012027 |
1.024615386 |
- |
0.512320016 |
- |
| 18 |
c.8212G>A |
8440G>A |
p.A2738T |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.082639742 |
1.024615386 |
- |
1.109289337 |
- |
| 18 |
c.8215G>A |
8443G>A |
p.V2739I |
0.083262903 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
0.89 |
1.840798594 |
1.792507344 |
- |
2.936684049 |
- |
| 18 |
c.8229_8243del |
8457del15 |
- |
0.918470115 |
3 - Uncertain |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.81 |
- |
- |
2.397581004 |
1.102157073 |
- |
2.64251086 |
- |
| 18 |
c.8243G>A |
8471G>A |
p.G2748D |
1.0000 |
5 - Definitely pathogenic |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.01 |
0.00 |
0.81 |
6.761 |
- |
257.04 |
1.445 |
- |
2511.19 |
- |
|
|
| DNA change |
|
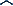
 | |
|
|
|
|
| Key Observational Reference |
 |
 | |
|
| Missense analysis Prior P |
 |
 | |
|
|
|
|
|
|
|
|
|
|
| 18 |
c.8249_8251del |
8477del3 |
- |
- |
- |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.81 |
- |
1.007512 |
0.580175601 |
1.075678821 |
- |
0.628770715 |
- |
| 18 |
c.8254A>T |
8482A>T |
p.I2752F |
0.010754887 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.334834862 |
1.049836689 |
- |
0.351521923 |
- |
| 18 |
c.8255T>A |
8483T>A |
p.I2752N |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
1.5268 |
- |
- |
- |
- |
1.5268 |
- |
| 18 |
c.8308G>A
(Reported 2 times) |
8536G>A |
p.A2770T |
0.0150 |
2 - Likely not pathogenic or of little clinical significance |
Chenevix-Trench, G. et al., Cancer Research 66: 2019-2027, 2006. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.03 |
0.01 |
0.00 |
0.03 |
0.41 |
1.2 |
- |
1 |
- |
0.492 |
- |
| 18 |
c.8309C>A |
8537C>A |
p.A2770D |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.907743458 |
1.049836689 |
- |
0.952982386 |
- |
| 18 |
c.8323A>G |
8551A>G |
p.M2775V |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.623051427 |
1.024615386 |
- |
0.638388078 |
- |
| 18 |
c.8324T>C |
8552T>C |
p.M2775T |
0.014182651 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.443087685 |
1.049836689 |
- |
0.465169708 |
- |
| 18 |
c.8331+1G>A |
IVS18+1G>A |
- |
0.989036504 |
4 - Likely Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
- |
- |
2.723026628 |
1.024615386 |
- |
2.790054978 |
- |
| 18 |
c.8331+1G>T |
IVS18+1G>T |
- |
0.985310009 |
4 - Likely Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
1.172 |
1.77 |
- |
- |
- |
2.07444 |
- |
| 18 |
c.8331+2T>C |
IVS18+2T>C |
- |
0.85934908 |
3 - Uncertain |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
0.567967269 |
0.66 |
0.491981128 |
1.024615386 |
- |
0.188962907 |
- |
| 18 |
c.8331+16C>G |
IVS18+16C>G |
- |
0.001201696 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.02 |
- |
0.184275908 |
- |
- |
0.319922071 |
0.05895393 |
- |
| 18 |
c.8332-13T>C |
IVS18-13T>C |
- |
- |
- |
Parsons et al 2019 |
- |
- |
0.34 |
0.02 |
0.34 |
- |
- |
1.499287799 |
1.024615386 |
- |
1.536193346 |
- |
| 19 |
c.8342A>T |
8570A>T |
p.N2781I |
0.627059345 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
- |
- |
0.366652694 |
1.075678821 |
- |
0.394400537 |
- |
| 19 |
c.8350C>T |
8578C>T |
p.R2784W |
0.8365 |
3 - Uncertain |
Farrugia DJ et al., Cancer Research 68:3523-3531, 2008. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.81 |
0.01 |
0.00 |
0.81 |
- |
- |
0.851 |
1.41 |
- |
1.19991 |
- |
| 19 |
c.8351G>A |
8579G>A |
p.R2784Q |
1.69404E-05 |
1 - Not pathogenic or of no clinical significance |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
0.000106872 |
1.134894587 |
1.480815593 |
0.048590003 |
- |
0.00000872701 |
Two functional assays report that this variant has a complete functional impact (Mesman et al 2018, Hart et al 2018). |
| 19 |
c.8356G>A |
8584G>A |
p.A2786T |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
1.06 |
0.580175601 |
1.075678821 |
- |
0.661527563 |
- |
| 19 |
c.8356G>C |
8584G>C |
p.A2786P |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
1.353124142 |
1.049836689 |
- |
1.420559368 |
- |
| 19 |
c.8359C>T |
8587C>T |
p.R2787C |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.561876016 |
1.18556701 |
- |
0.666141669 |
- |
| 19 |
c.8360G>A |
8588G>A |
p.R2787H |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
1.9948 |
0.6549 |
1.24792346 |
1.049836689 |
- |
1.711528144 |
- |
| 19 |
c.8362T>C |
8590T>C |
p.W2788R |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.3 |
- |
- |
0.731690901 |
1.024615386 |
- |
0.749701754 |
- |
| 19 |
c.8363G>C |
8591G>C |
p.W2788S |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
- |
1.066689585 |
1.024615386 |
- |
1.09294656 |
- |
| 19 |
c.8386C>T |
8614C>T |
p.P2796S |
0.003798709 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
0.37 |
0.309781167 |
1.075678821 |
- |
0.123293265 |
- |
| 19 |
c.8417C>T |
8645C>T |
p.S2806L |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
0.8672 |
- |
- |
- |
- |
0.8672 |
- |
| 19 |
c.8421G>T |
8649G>T |
p.S2807S |
- |
- |
Parsons et al 2019 |
- |
0.02 |
- |
0.02 |
0.02 |
- |
- |
- |
1.025 |
- |
1.025 |
- |
| 19 |
c.8428A>G |
8656A>G |
p.S2810G |
0.003949435 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.113527132 |
1.129287087 |
- |
0.128204724 |
- |
|
|
| DNA change |
|
 | |
|
|
|
|
| Key Observational Reference |
 |
 | |
|
| Missense analysis Prior P |
 |
 | |
|
|
|
|
|
|
|
|
|
|
| 19 |
c.8432A>G |
8660A>G |
p.D2811G |
0.013123908 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.399731529 |
1.075678821 |
- |
0.429982739 |
- |
| 19 |
c.8438G>A |
8666G>A |
p.G2813E |
- |
- |
Parsons et al 2019 |
- |
0.81 |
- |
0.02 |
0.81 |
- |
- |
0.766838466 |
1.049836689 |
- |
0.805055155 |
- |
| 19 |
c.8444T>G |
8672T>G |
p.V2815G |
0.487440238 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
- |
0.478135484 |
1.024615386 |
- |
0.489904974 |
- |
| 19 |
c.8455G>A |
8683G>A |
p.D2819N |
- |
- |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
- |
- |
0.70265866 |
1.024615386 |
- |
0.719954874 |
- |
| 19 |
c.8464_8466del |
8692del3 |
- |
0.483468071 |
3 - Uncertain |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.66 |
- |
- |
0.470592211 |
1.024615386 |
- |
0.482176019 |
- |
| 19 |
c.8486A>G |
8714A>G |
p.Q2829R |
0.933371967 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.03 |
0.34 |
0.02 |
0.34 |
14.29500979 |
1.902297 |
- |
- |
- |
27.19335423 |
- |
| 19 |
c.8487G>A |
8715G>A |
p.Q2829Q |
- |
- |
Parsons et al 2019 |
- |
0.02 |
0.34 |
0.02 |
0.34 |
0.9985 |
- |
1.106616322 |
1.049836689 |
- |
1.160023766 |
- |
| 19 |
c.8487+1G>A |
IVS19+1G>A |
- |
0.999 |
5 - Definitely pathogenic |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Vallee MP et al., Human Mutation 37: 627-639, 2016 |
- |
0.97 |
0.00 |
0.97 |
1 |
1 |
13.18 |
1.59 |
- |
20.89 |
- |
| 19 |
c.8487+4T>C |
IVS19+4T>C |
- |
0.00975981 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
- |
0.04 |
0.02 |
0.04 |
- |
- |
0.230861316 |
1.024615386 |
- |
0.236544056 |
- |
| 19 |
c.8488-19G>A |
IVS19-19G>A |
- |
- |
- |
Parsons et al 2019 |
- |
- |
0.34 |
0.02 |
0.34 |
- |
- |
0.845804261 |
1.049836689 |
- |
0.887956344 |
- |
| 19 |
c.8488-1G>A |
IVS19-1G>A |
- |
0.992712204 |
5 - Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
- |
- |
3.730545463 |
1.129287087 |
- |
4.212856817 |
- |
| 20 |
c.8503T>C |
8731T>C |
p.S2835P |
0.000863314 |
1 - Not pathogenic or of no clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
0.10565456 |
1.06 |
0.249459481 |
- |
- |
0.027937924 |
- |
| 20 |
c.8507C>G |
8735C>G |
p.S2836C |
0.076378791 |
3 - Uncertain |
Parsons et al 2019 |
- |
0.29 |
- |
0.02 |
0.29 |
0.191 |
1.06 |
- |
- |
- |
0.20246 |
- |
| 20 |
c.8515T>C |
8743T>C |
p.Y2839H |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.963486766 |
1.024615386 |
- |
0.987203364 |
- |
| 20 |
c.8518A>G |
8746A>G |
p.I2840V |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.845804261 |
1.102157073 |
- |
0.932209148 |
- |
| 20 |
c.8525G>A |
8753G>A |
p.R2842H |
0.00026 |
1 - Not pathogenic or of no clinical significance |
Easton DF et al., Am J Hum Genet, 81: 873-883, 2007. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.29 |
0.01 |
0.00 |
0.29 |
0.07586 |
- |
0.14125 |
0.05888 |
- |
0.00063 |
- |
| 20 |
c.8567A>C |
8795A>C |
p.E2856A |
0.0398 |
2 - Likely not pathogenic or of little clinical significance |
Chenevix-Trench, G. et al., Cancer Research 66: 2019-2027, 2006. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.03 |
0.01 |
0.00 |
0.03 |
1.05 |
1.2 |
- |
1.064 |
- |
1.34064 |
- |
| 20 |
c.8572C>A |
8800C>A |
p.Q2858K |
0.005118984 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.150945301 |
1.102157073 |
- |
0.166365431 |
- |
| 20 |
c.8573A>G
(Reported 2 times) |
8801A>G |
p.Q2858R |
0.0187 |
2 - Likely not pathogenic or of little clinical significance |
Spurdle AB et al., J Clin Oncol, 26: 1657-1663, 2008. |
Tavtigian, S.V. et al., Journal of Medical Genetics 43: 295-305, 2006. |
0.03 |
0.01 |
0.00 |
0.03 |
0.938 |
0.5 |
- |
1.3158 |
- |
0.61711 |
- |
| 20 |
c.8591C>T |
8819C>T |
p.A2864V |
- |
- |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.975545041 |
1.049836689 |
- |
1.024162976 |
- |
| 20 |
c.8593T>G
(Reported 2 times) |
8821T>G |
p.L2865V |
0.2443 |
3 - Uncertain |
Farrugia DJ et al., Cancer Research 68:3523-3531, 2008. |
Tavtigian et al., Human Mutation 29: 1342-1354, 2008. |
0.29 |
0.01 |
0.00 |
0.29 |
- |
- |
0.776 |
1.02 |
- |
0.79152 |
- |
| 20 |
c.8599A>C |
8827A>C |
p.T2867P |
0.007877847 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
0.03 |
- |
0.02 |
0.03 |
- |
- |
0.244551939 |
1.049836689 |
- |
0.256739597 |
- |
| 20 |
c.8618T>G |
8846T>G |
p.F2873C |
- |
- |
Parsons et al 2019 |
- |
0.66 |
- |
0.02 |
0.66 |
- |
- |
1.480743853 |
1.049836689 |
- |
1.554539223 |
- |
| 20 |
c.8632+1G>A |
IVS20+1G>A |
- |
0.99809348 |
5 - Pathogenic |
Parsons et al 2019 |
- |
- |
0.97 |
0.02 |
0.97 |
21.9304 |
0.7383 |
- |
- |
- |
16.19121432 |
- |
| 20 |
c.8632+15A>G |
IVS20+15A>G |
- |
0.003870674 |
2 - Likely not pathogenic or of little clinical significance |
Parsons et al 2019 |
- |
- |
- |
0.02 |
0.02 |
0.1904 |
- |
- |
- |
- |
0.1904 |
- |
